Methods in biostatistics and bioinformatics
Christian Dina, Alban Gaignard, Pierre Lindenbaum, Richard Redon
Investigators : Christian Dina, Alban Gaignard, Pierre Lindenbaum, Richard Redon
Support staff : Floriane Simonet
Because most of our projects exploit massive multi-source datasets and multiple variables simultaneously we plan to further develop and apply state-of-the-art methods in bioinformatics & biostatistics to better understand, prevent and treat cardiovascular disease. Our objective is to capitalize on our recent methodological developments and expansion of artificial intelligence methods. Our ambition is to provide each research theme with the most innovative methodological tools for disentangling the genetic architecture of disease risk and explore new approaches for identifying biological and evolutionary mechanisms using genomic data.
First, we aim at expanding multi-omics studies by performing association studies on the whole spectrum of allele frequencies on patients with cardiac valve diseases, cardiac arrhythmias and intracranial aneurysms. More specifically, we combine innovative statistical tools with methods based on machine learning, knowledge representation and logical reasoning. We build flexible frameworks to allow efficient gene and pathways discovery from heterogeneous data. While the traditional approach is first applying statistical methods and combine marker or set of associated markers statistics with existing information (cell-line chromatin accessibility, chromatin conformation neighbor gene pathways…), this approach maybe hampered by moderate sample size for the diseases we are studying. We propose a framework to apply statistical methods that will integrate prior functional probabilities from annotation data in order to improve our power.
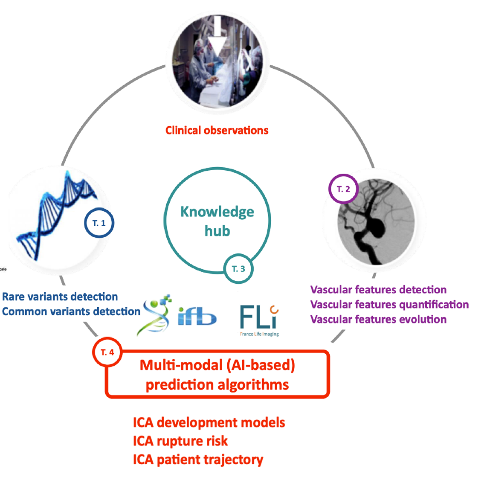
Second, we integrate large imaging datasets with Omics data to discover key biological pathways involved in cardiovascular disease and propose novel diagnosis or prognosis approaches. More specifically we work at proposing FAIR (Findable, Accessible, Interoperable, Reusable) algorithms and datasets for better integrating, exploiting and modeling massive and heterogeneous biomedical data. We will primarily focus on better predicting the formation and development of intracranial aneurysms through the additional use of imaging data. This approach will then be translated to rare diseases such as Brugada syndrome.
Third, we intend to combine multi-omic data from cohort and population datasets to improve our knowledge on the complex architecture underlying cardiac arrhythmia and thus improve disease risk assessment and therapeutic strategies.
We will then integrate genetic variants associated to disease in single metrics, considering genetic evolution and shared genetic architecture. We will use large biobanks and Electronic Health Records together with PRS to explore endo-phenotypes and further elaborate on genetic architecture and on pathophysiological mechanisms.
Alves I, Giemza J, Blum MGB, Bernhardsson C, Chatel S, Karakachoff M, Saint Pierre A, Herzig AF, Olaso R, Monteil M, Gallien V, Cabot E, Svensson E, Bacq D, Baron E, Berthelier C, Besse C, Blanché H, Bocher O, Boland A, Bonnaud S, Charpentier E, Dandine-Roulland C, Férec C, Fruchet C, Lecointe S, Le Floch E, Ludwig TE, Marenne G, Meyer V, Quellery E, Racimo F, Rouault K, Sandron F, Schott J-J, Velo-Suarez L, Violleau J, Willerslev E, Coativy Y, Jézéquel M, Le Bris D, Nicolas C, Pailler Y, Goldberg M, Zins M, Le Marec H, Jakobsson M, Darlu P, Génin E, Deleuze J-F, Redon R, Dina C.
Nat Commun 2024;15:6710.
The genetic history of France.
Saint Pierre A, Giemza J, Alves I, Karakachoff M, Gaudin M, Amouyel P, Dartigues J-F, Tzourio C, Monteil M, Galan P, Hercberg S, Mathieson I, Redon R, Génin E, Dina C.
Eur J Hum Genet 2020;doi:10.1038/s41431-020-0584-1.
Findable and Reusable Workflow Data Products: A Genomic Workflow Case Study.
Gaignard A, Skaf-Molli H, Belhajjame K.
Semantic Web journal, 2019 10.3233/SW-200374
bioalcidae, samjs and vcffilterjs: object-oriented formatters and filters for bioinformatics files.
Lindenbaum P, Redon R.
Bioinformatics 2018;34:1224–1225.
DoEstRare: A statistical test to identify local enrichments in rare genomic variants associated with disease.
Persyn E, Karakachoff M, Le Scouarnec S, Le Clézio C, Campion D, Consortium FE, Schott J-J, Redon R, Bellanger L, Dina C.
PLoS ONE 2017;12:e0179364.
Funding
- Inserm
- NExT